By Dr. Anwesha Ghosh, PhD, Manager – Scientific Affairs and Communications Specialist
Genetic variation can range from changes at the level of single bases to whole-chromosomal aneuploidies. Structural variations (SVs) refer to a large alterations in chromosomal structure, typically encompassing larger than 1 Kbp of DNA. SVs include both balanced changes, such as inversions and some forms of translocations, as well as those that alter DNA copy number through duplications and deletions of chromosomal segments. SVs account for 25% of protein truncating mutations and are 3 times more likely to associate with a genome-wide association study (GWAS) signal than single nucleotide variants (SNVs). SVs contribute to all classes of genetic disease: sporadic development syndromes, Mendelian diseases, complex disorders and infectious diseases, as well as health-related metabolic phenotypes.
While next generation sequencing (NGS) has enabled extensive characterization of SNVs in the human genome, the short length of sequencing reads it employs impairs its ability to provide insight into larger genomic changes like SVs. Conventional methods to detect SVs include karyotyping, fluorescence in situ hybridization (FISH) and chromosomal microarray analysis (CMA). While karyotyping is cost effective, it suffers from numerous drawbacks, including low resolution, high labor and time consumption, requirement for cell culture, and subjectivity in interpretation. FISH, while not requiring cell culture and significantly improving resolution, is a targeted approach that cannot provide genome-wide information. While CMA can overcome these limitations, it cannot detect certain classes of SVs, such as balanced translocations or inversions, expansions of repeat regions, or low-level mosaicism.
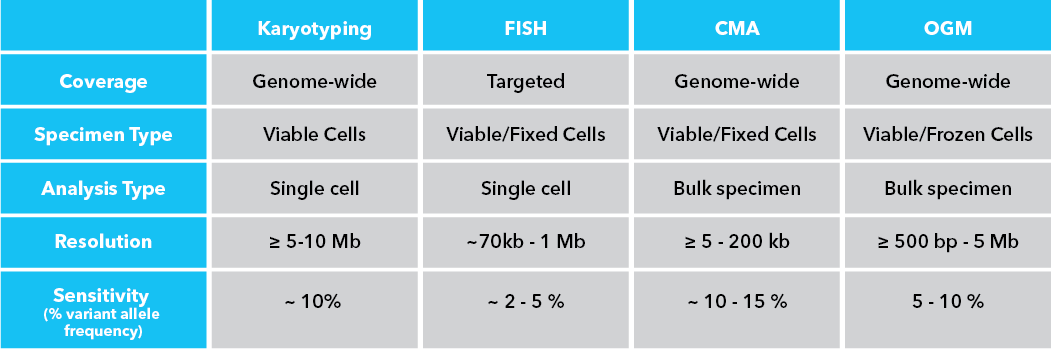
Optical genome mapping (OGM) offers a solution to these pitfalls by essentially combining the genome-wide scope of karyotyping with the visualization principle of FISH into a single workflow that provides data at the highest resolution reported for the field of cytogenetics at 500 bp, while successfully detecting all classes of SVs. A comparison of the requirements, scope, and performance of karyotyping, FISH, CMA and OGM is provided in Table 1.
Table 1
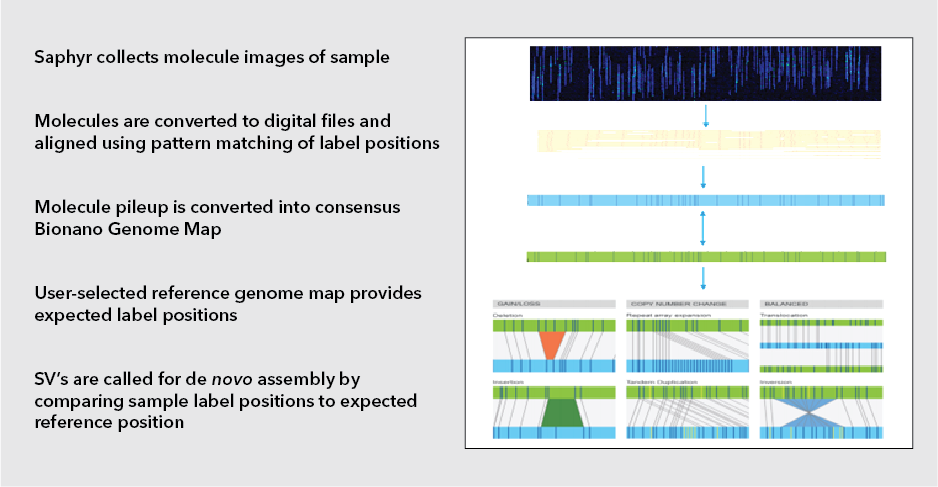
OGM relies on the isolation of intact, ultra-high molecular weight (UHMW) DNA using an extraction protocol specifically designed to minimize the shearing forces generated by typical standard column-based extraction methods. This yields DNA fragments of ~150 Kb to a few Mb in size. This DNA is then fluorescently labeled via covalent modification at CTTAAG hexamer motifs which occur throughout the genome at a frequency of ~14-17 per 100kb in sequence specific patterns. The labeled DNA is loaded on silicon microfluidic chips containing thousands of parallel nanochannels in which individual DNA molecules are linearized, imaged, and digitized. Each DNA fragment bears a distinct spacing and pattern of the hexamer labels (also known as the label profile), which are subsequently grouped and aligned based on label profile matching to produce consensus maps. These maps are then compared in silico to the expected labeling profile of a reference genome. DNA containing structural variations will display a labeling profile that differs from the reference genome at the location of the variation. The type of structural variation present can be determined based on the nature of the altered labeling profile (Fig 1). Bionano’s Saphyr system is currently the best in class for OGM.
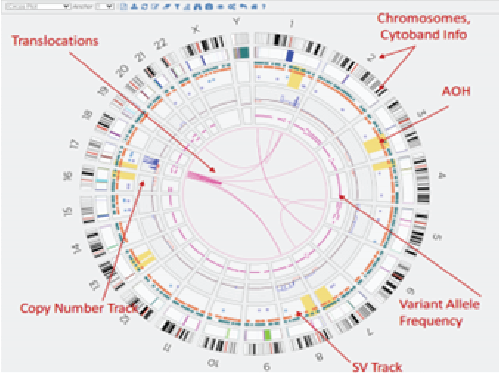
SV calling can be performed using Bionano pipelines such as annotated de novo assembly for somatic variations or the annotated rare variant pipeline for germline variations. A snapshot of all SVs detected in the genome in any given experiment can be viewed in a circos plot, while closer inspection of specific regions can be performed in the genome map view (Figure 2).
Bionano EnFocus pipelines have also been developed for targeted detection of specific genomic variations known to be found in diseases like facioscapulohumeral muscular dystrophy (FSDH) and Fragile X syndrome.
Circos Plot
Genome Map View
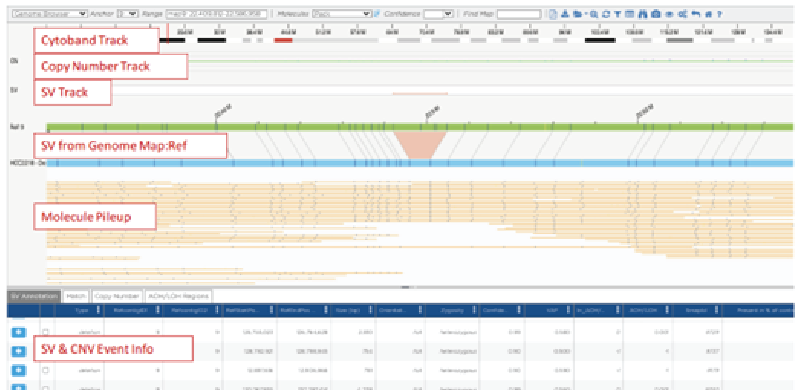
Figure 2: Circos plots and genome map views of data obtained by OGM
Currently, MedGenome offers Bionano’s EnFocus solution for detection of FSHD, which is the third most common inherited skeletal muscle disease. FSHD is associated with contraction of the D4Z4 microsatellite repeat regions within the sub-telomeric region of chromosome 4q35. Normal individuals harbor 11-100 such repeats, while afflicted individuals harbor less than 10 repeats. The current standard of care to confirm an FSHD diagnosis is mainly through a Southern blot assay. However, Southern blots are time and labor intensive with a greater scope for human error, which are drawbacks that can be overcome with the OGM approach.
OGM has applications in the fields of hematological malignancies, solid tumor research, constitutional genetic disorders and quality control for cell and gene therapy such as CAR-T immune cell therapy. As a precision medicine tool for hemato-oncology, it can identify classical actionable fusions (such as the BCR-ABL1 fusion in acute lymphoblastic leukemia) and could enable patient stratification based on signature SVs associated with biological phenomena underlying specific therapeutic sensitivities, such as high replication stress. It can easily identify multiple complex rearrangements within a single patient in challenging cases, thereby precluding the need to perform multiple conventional techniques. It can also identify novel variations, such as those that are missed by sequencing because they are too long or high in GC content. Additionally, it unlocks the potential to characterize SV landscape of solid tumors, which had remained largely unexplored due to the challenges of karyotyping. Finally, when combined with NGS, OGM can not only provide a more comprehensive understanding of cancers, but also aid the diagnosis of rare monogenic diseases.
#Optical genome mapping (OGM), #OGM, #Structural variations, #ultra-high molecular weight (UHMW) DNA, #Bionano’s Saphyr system, #Facioscapulohumeral muscular dystrophy (FSDH), #Fragile X syndrome
 US
US IN
IN

