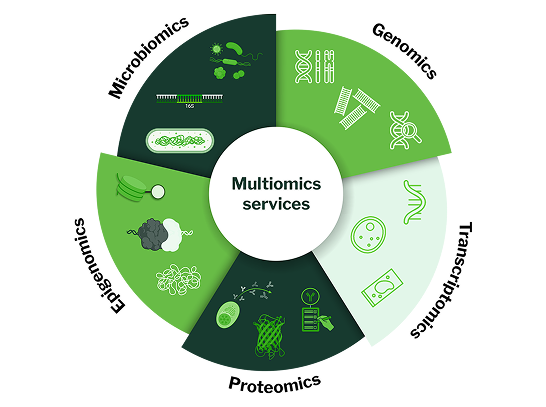
Signios Bio, the US arm of MedGenome—a global leader in genetic testing and genomics research—is a multiomics and bioinformatics company dedicated to decoding complex disease biology. We integrate data across genomics, transcriptomics, proteomics, epigenomics, metabolomics, and microbiomics to generate comprehensive insights. Our AI-powered bioinformatics platform uncovers hidden patterns in biological data, accelerating the development of new therapies and diagnostics.
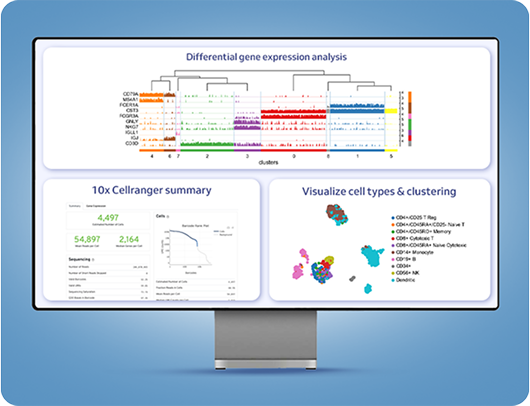
Sequencing capabilities allow for in-depth investigation into tumor genomics, tumor microenvironments, and molecular pathways driving cancer progression
Immune profiling of single cells assist in studying immune system dynamics, immune responses, and the mechanisms of immune evasion in cancer
Whole genome and exome sequencing to enable discovery of novel genetic mutations and understanding the genetic basis of rare disorders
Sequencing capabilities allow for in-depth investigation into tumor genomics, tumor microenvironments, and molecular pathways driving cancer progression
RWE supports epidemiological studies, population health research, and the development of inclusive therapies that address global health disparities
Multiomics aids in identifying biomarkers for drug efficacy, safety, and dosage optimization, advancing personalized treatment strategies in various therapeutic areas
Single-cells sequenced
Unique variants identified
Transcriptomes sequenced
Peer-reviewed publications
Citations of our work
DNA sequencing assays run
