By Vinay CG, Associate Director, Content & Communications and MedGenome Scientific Affairs
The human immune response can be divided into two components: Innate and Adaptive. Innate immune response involves classic primitive reaction through cellular and humoral mechanisms. It’s a first line of defence and can comprise a host of cells such as neutrophils, macrophages, and mast cells which kills the invading pathogens while the humoral response can be through enzymes such as Lysozyme that can kill harmful microorganisms.
The most effective component of the Immune system is the Adaptive immunity. Also, known as Acquired immunity – it is a highly advanced evolutionary system which recognizes, identifies pathogens, and tailors a specific response towards them. This system essentially involves Lymphocytes. Each lymphocyte expresses a receptor on its surface that can specifically bind to a particular antigen.
Even the adaptive immunity has two arms to it the cellular and the humoral. The cellular arm comprises of T Lymphocytes (T cells) which help in eliminating pathogens through various mechanisms while the humoral arm involves a subset of lymphocytes called B Lymphocytes (B cells).
The T cells recognize antigen through T-cell antigen receptor (TCR) and the B cells recognize intact antigens through immunoglobulins (antibodies). B cells too have receptors termed as B-cell antigen receptors (BCR).
Profiling the TCRs and BCRs (Figure 1) holds a great promise in understanding mechanisms that can provide extremely useful insights in developing new therapeutics and addressing critical research questions in the areas of translational immunology, immunotherapy, autoimmune disorders, and transplant research [1,2].
TCR Profiling
TCRs are made up of heterodimers α/β (TCR2) or γ/δ (TCR1) chains [3]. These chains are encoded by 4 different gene loci namely V (variable), D (diversity), J (joining) and C (constant). A typical T cell will either express an α/β or a γ/δ receptor [4]. Similarly, BCRs are made up of heavy and light chains [4].
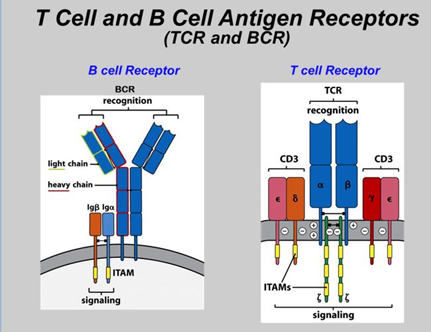
The TCRs are diverse in nature owing to the extensive recombination between different V, D, and J gene segments. These rearrangements can lead to an incredibly (109-1010 sequences) large spectrum of complementary-determining region 3 (CDR3) [1]. CDR3 is critical in binding to their specific antigens. When a TCR expands by binding to an antigen there is a selective expansion in the CDR3 region of the repertoire resulting in a clonotype. With effective NGS based T cell receptor sequencing methodologies and assay it is now possible to identify all such clonotypes in a diverse repertoire of TCRs. This allows researchers to analyze the TCR repertoire in patients and aid in predicting better treatment outcomes.
TCR repertoire sequencing can provide deeper understanding of the clonal diversity, richness and evenness of the T Cell clonal population which in turn can help in identifying biomarkers for better immunotherapy responses [5].
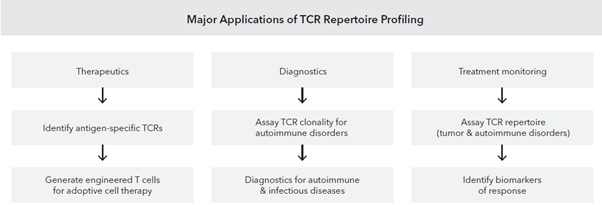
At MedGenome, we have standardized methodologies for bulk TCR sequencing where we have obtained expertise on working with diverse input types to obtain TCR α/β, γ/δ clonotypes (for bulk – cells, RNA and FFPE tissues) and TCR α/β clonotypes from single-cell inputs [1]. Additionally, our powerful bioinformatics analysis pipeline provides broader insights such as full length clonotype sequences, V-J usage summaries, CDR3 length distribution, and shared clonotype analysis (Figure 2).
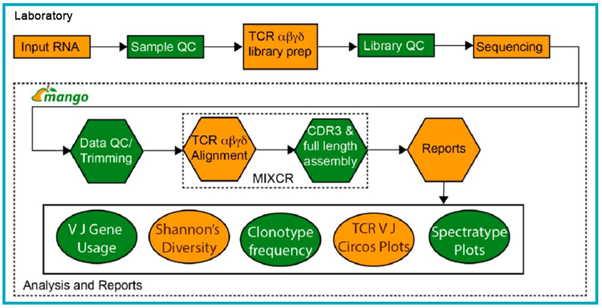
Starting with total RNA either obtained from client or prepared in-house, sample QC is performed using the Agilent’s TapeStation or Fragment analyzer. If samples meet the QC criteria, then they are processed using the SMARTer TCR α/β Profiling kit or a modified protocol for γ/δ TCR profiling. After library preparation, QC is performed using Agilent’s TapeStation or Fragment analyzer, and sequencing is performed on Illumina platform. Data generated is demultiplexed and FastQC is performed, and after trimming, MiXCR software is used for alignment of reads to the TCR clonotypes, and the final CDR3 and full length clonotypes are assembled. Advanced analyses outputs can also be generated to compare VJ gene usage across samples, and determine Shannon’s diversity and frequency changes in the repertoire across samples.
BCR Profiling
The B cells too play an important role in immune response. However, they also play havoc when they go wrong causing several B-cell mediated diseases such as cancer mostly known and noted type being B-cell malignancies such as non-Hodgkin’s lymphoma and Hodgkin’s lymphoma, autoimmune diseases such as multiple sclerosis, rheumatoid arthritis and systemic lupus erythematosus [6,7] As a B cell matures several recombination of immunoglobulin genes occur owing to an event called class switch recombination (CSR) which can lead to diversification of the B-cell repertoires [8]. Further, B-cell receptors can undergo variations when it binds to a certain antigen. through V(D)J recombination, class switch recombination, and somatic hypermutation that can cause DNA breaks triggering cancer development [8].
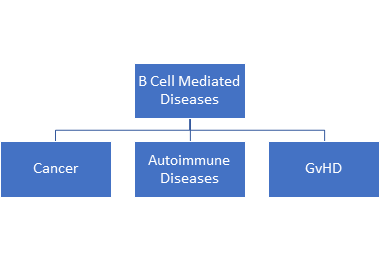
Therefore, BCR sequencing gives an insight into the various interplay between B-cell differentiation, BCR somatic hypermutation, class switching, and antigen specificity.
Major Applications of BCR Repertoire Profiling
At MedGenome, we have also developed and standardized, HitMab (High-throughput antibody discovery using single cell sequencing), a streamlined workflow for antibody discovery using high-throughput single-cell B-cell receptor sequencing (scBCR-seq). HitMab allows us to obtain accurately paired full-length variable regions in a massively parallel fashion. With HitMab’s single cell BCR sequencing it is possible for researchers to speed up discovery of thousands of antibodies at a much lesser time compared to the traditional Hybridoma Technology.
Want to know more about our exciting TCR/BCR Solutions?
Get in touch with our experienced and seasoned scientific team to understand how our unique immune repertoire solutions can provide deeper insights to your research projects. You can also email us at research@medgenome.com for any queries and further details.
References
- 1. MedGenome TCR White Papers, https://research.medgenome.com/medgenome-documents/
- 2. 10x Genomics, Inc. LIT000017 Rev C Chromium Single Cell Immune Profiling Product Sheet
- 3. Transplant International 2019; 32: 1111–1123
- 4. David Male, Immunology an illustrated outline, 4th Edition, 2004, Elsevier
- 5. https://www.thermofisher.com/in/en/home/life-science/sequencing/sequencing-learning-center/next-generation-sequencing-information/immuno-oncology-research/why-immune-repertoire-matters.html
- 6. Bite-sized immunology by British Society of Immunology.
- 7. Bashford-Rogers, R.J.M., Bergamaschi, L., McKinney, E.F. et al. Analysis of the B cell receptor repertoire in six immune-mediated diseases. Nature 574, 122–126 (2019). https://doi.org/10.1038/s41586-019-1595-3
- 8. Illumina Immuno-oncology overview publication.
#t cell receptor sequencing, #tcr repertoire sequencing, #t cell repertoire sequencing, #bcr sequencing, #TCR repertoire, #NGS based t cell receptor sequencing
 US
US IN
IN

