MedGenome specializes in processing challenging samples including FFPE and low-quality input material. Our analysis includes comprehensive report with rich visualizations identifying all types of relevant DNA and RNA variants.

Identifying and prioritizing the disease causing variants from thousands of variants that are called during a whole genome and exome sequencing analysis is a time consuming and manual task. Pathogenicity based ranking of variants greatly improves the speed of report generation, in turn increasing the diagnostic yield.

Single-cell transcriptomics has revolutionized genomics and is now an integral part of therapeutics and diagnostics research. Single cell RNA sequencing (scRNA-seq) enables the analysis of gene expression at single cell resolution via droplet-based cell capture methods that rely on microfluidic instruments such as those developed by 10x genomics.

Single cell genomic approaches can provide valuable insights into the complexity and heterogeneity of the cell types in the context of a tissue or tumor. However, challenges with the preparation of single cell suspensions, good cell viability and efficiently capturing diverse cell types in a mix via appropriate cell capture methods can override the utility of the approaches.
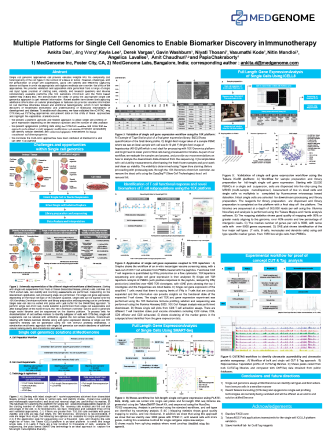
T cell immunity provides significant therapeutic benefit to cancer patients treated with checkpoint inhibitors, however a very small fraction of patients typically respond to checkpoint inhibitors, and a smaller fraction of them have any long-term benefit. This can be attributed to the lack of prognostic and predictive biomarkers.

While Genome wide association studies can shed light on the significance of variants in susceptibility to a disease or allow to stratify patients for specific therapeutic modalities, often variants that are rare and could be of significance are not identified in these studies.

Medical big data analytics has applications in clinical decision, predictive/ prognostic modelling of disease progression, disease surveillance, public health and research.

Somatic mutations have been found to be a rich source of potential cancer vaccines (which have shown promise in treating late stage cancers) with minimal T cell tolerance. MedGenome has built a proprietary cancer vaccine prediction platform, OncoPeptVAC using a combination of features that include TCR binding, human leukocyte antigen (HLA) binding, gene expression and proteasomal processing.

Papia Chakraborty3, Snigdha Majumder1, Rakshit Shah2, Jisha Elias1,2, Vasumathi Kode3, Yogesh Mistry2, Coral Karunakaran1, Priyanka Shah1, Malini Manoharan1, Bharti Mittal1, Sakthivel Murugan SM1, Lakshmi Mahadevan1, Ravi Gupta1, Amitabha Chaudhuri1,3 ** and Arati Khanna-Gupta1**
The MedGenome team has identified a germline mutation in an MMR pathway (the DNA mismatch repair pathway) gene – MLH1. Mutations in the MMR pathway genes have been known to be associated with Lynch syndrome wherein patients have a 70-80% lifetime risk of developing colorectal cancer (CRC).
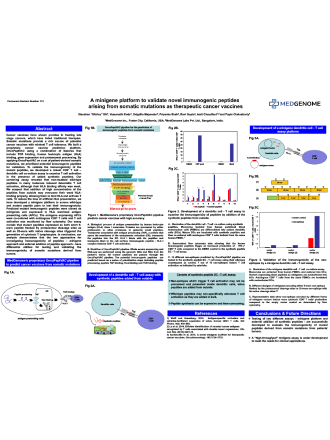
Neoantigens, derived from somatic mutations are prime candidates for cancer vaccines. Currently, the available T-cell neoepitope prioritization pipelines rely primarily on two attributes – the class-I HLA-binding affinity of the mutant peptide compared to the wild-type counterpart, and the level of expression of the mutated gene in tumor cells.

Ashwini Patil, MS1, Ravi Gupta, PhD1, Nitin Mandloi, MS1, Kiran V. Paul, MS1, Priyanka Shah, PhD1, Malini Manoharan, PhD1, Rohit Gupta, PhD1, and Amitabha Chaudhuri, PhD1
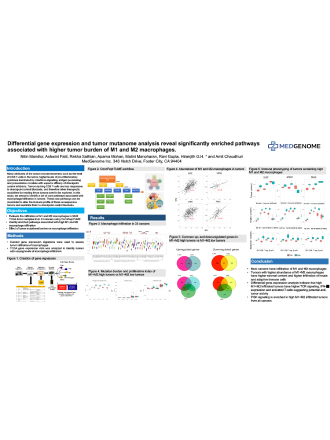
MedGenome conducted a study where TCGA data containing 9640 tumors from 33 different cancers was analyzed using its proprietary tumor microenvironment analysis platform OncoPeptTUMETM. It was observed that observed that CD8 T-cell content of tumors varies significantly from cancer to cancer, with a large proportion of tumors containing low CD8 T-cell infiltrate.

MedGenome conducted a study in which TCGA data was analyzed to investigate the impact of CD8 T-cell infiltration on disease outcome. The analysis indicated that CD8 T-cell infiltration predicts favorable survival in certain cancers, whereas in other cancers it has no effect.

With the knowledge that tumors lacking CD8 T cells are less responsive to checkpoint control blockade, MedGenome’s scientists have chosen to study a set of core pathways associated with the absence or presence of specific immune cell types in tumors, which can be modulated to alter the immune profile of these unresponsive tumors and sensitize them to checkpoint control blockade.

In the backdrop of the remarkable success checkpoint control inhibitors have shown in treating a variety of different cancers, this study focused on deeper assessment of the tumor and its microenvironment at the genetic and phenotypic level. Data from recent clinical trials have unequivocally established that the tumor microenvironment significantly impacts the efficacy of immune-oncology drugs.
With only a few studies having analyzed the interaction between granulocytic and monocytic myeloid derived suppressor cells in human cancers, MedGenome’s scientists have chosen to investigate their immune suppressive effect on the tumor microenvironment in this study.

A critical phase in the development of cancer is the conversion of growth-suppressive normal tissue microenvironment into a growth-promoting tumor microenvironment. MedGenome conducted a study to interrogate the tumor epithelial and the stromal compartments in a cohort of tongue and buccal cancers, using NGS sequencing.

Large scale sequencing of cancer genomes have revealed several mutations that remain uncharacterized, of which only few mutations are tested for activating or loss of function. MedGenome conducted a study which utilized in-silico method to characterize 4096 mutations in 190 cancer census genes spread across 33 cancer groups.

It is known that tumor cells employ a variety of immune-evasive mechanisms. Of particular interest is the tumor-intrinsic mechanisms that prevent T-cells from infiltrating into the tumor microenvironment.

At the conference, MedGenome introduced OncoPept- its integrated platform that combines powerful analytics to analyze exome and RNA-seq data to quantitate the epithelial, stromal and immune cell infiltration, thereby producing a holistic view of the tumor and the tumor microenvironment.

Viswanathan S.1, Gupta R.1, Khanna-Gupta A.1, Chaudhuri A.2
The study titled “Whole Genome Sequencing data from the Wellderley study identifies rare variants in genes associated with diabetes and cardiomyopathy” involved the analysis of publicly available whole genome sequence data (WGS) of 454 healthy elderly Caucasian individuals from the Wellderly study.

Head and Neck squamous cell carcinoma (HNSCC) comprises of tumors arising from multiple sites of the upper aerodigestive tract. Cancers of buccal and tongue are highly prevalent worldwide and account for significant morbidity and mortality.

MedGenome presented results from a study that examined differential gene expression, that identified unique and shared vaccine candidates that can be used in combination therapies. The study involved analysis of published genomics data from over 400 buccal and tongue cancer samples to identify potential vaccine candidates using MedGenome’s proprietary analysis platform.

OncoMD Cancer Analytics Platform combines tumor mutation profiles, expression signatures, copy number variations, epigenetic alterations and drug sensitivity to create a holistic view of human cancer enabling discovery of new targets for therapy and prognosis.

OncoPeptTM is an integrated platform that combines powerful analytics to analyze exome and RNA-seq data with multistep prioritization to select cancer vaccine candidates as therapeutics. The platform generates neo-antigens from different types of genetic alterations such as SNVs, indels and fusion genes to generate a library of peptides that are automatically interrogated through the multiple prioritization steps.

This study reveals the functionality of OncoPept, MedGenome’s proprietary cancer immunotherapy platform, that predicts T-cell neo-epitopes from human and mouse cancers. Multiple pre-clinical and clinical studies have demonstrated the importance of the characterization of T-cell neo epitopes and its role in the development of cancer vaccines.

The poster depicts an oral cancer study to examine the molecular basis of region-specific differences by profiling four distinct oral cancer cohorts collected from the Northern, Eastern, Southern and Western regions of India. The cohorts in the study were such that it maximized the probability that all individuals shared similar environmental and lifestyle factors within a given region.

