By Dr. Lavanya Balakrishnan, Vinay C. G. and Michelle Balakrishnan Vierra, MedGenome Scientific Affairs
Single-cell sequencing offers unprecedented resolution for analyzing the unique molecular signatures of individual cells. By deconstructing the cellular landscape, it reveals hidden cell populations, tracks how cells differentiate, and identifies disease biomarkers. By dissecting the genomic, transcriptomic, and epigenetic landscape of single cells, researchers can now gain a deeper understanding of complex biological processes, disease mechanisms, and how patients respond to treatments. This method is essential for both basic and clinical research for unlocking the mysteries of cellular complexity and dynamic biological processes.
Single-cell sequencing: techniques and practical applications in cellular research
Single-cell sequencing utilizes a variety of techniques, with each assay tailored to target specific cellular information according to the investigator’s needs. Here’s an overview of key techniques and their applications:
Single-cell RNA sequencing (scRNA-seq)
scRNA-seq enables gene expression analysis at the single-cell level, revealing hidden cellular diversity. By profiling RNA molecules, it identifies new cell types and discovers disease-associated gene networks, facilitating biomarker discovery and treatment improvement. This technique accelerates drug development by enabling precise patient stratification and monitoring, leading the way to personalized medicine.
Case Example: Recent MedGenome-Supported Publication
A recent study by Pasqualina Colella et al. from Stanford University, published in Nature, employed scRNA-seq to reveal unexpected diversity in microglia-like cells within the brain. This finding shed light on how blood-derived cells can repopulate the central nervous system. Their work further demonstrates the therapeutic potential of stem cell transplants for progranulin-deficient neurodegenerative diseases1.
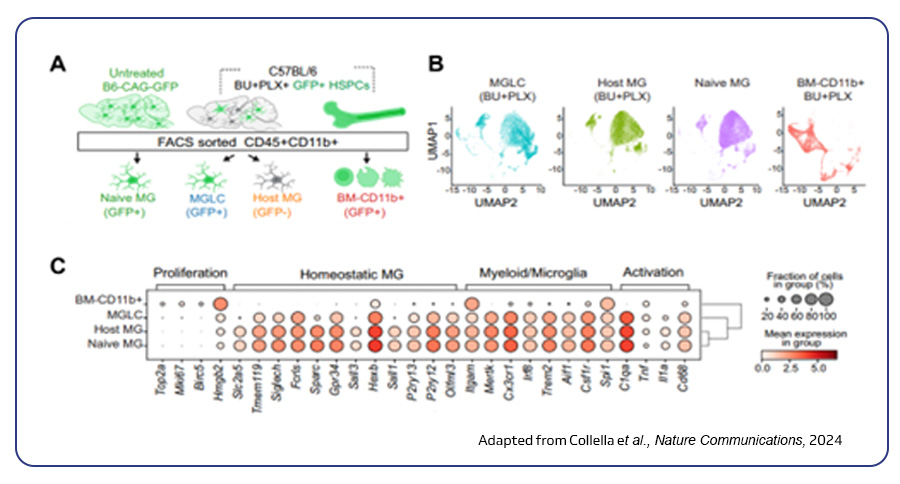
Single-cell DNA sequencing (scDNA-seq)
scDNA-seq is highly useful in understanding the genetic makeup of individual cells, identify driver mutations and copy number variations to reveal tumor heterogeneity and cancer evolution. One interesting publication by Ng et al. (2024) employed scDNA-seq to investigate the mechanisms leading to amplicons in esophageal adenocarcinoma, a cancer fueled by frequent gene amplifications. This advanced approach provided deeper insights into how amplified regions contribute to tumor evolution over time, offering valuable understanding of cancer progression2.
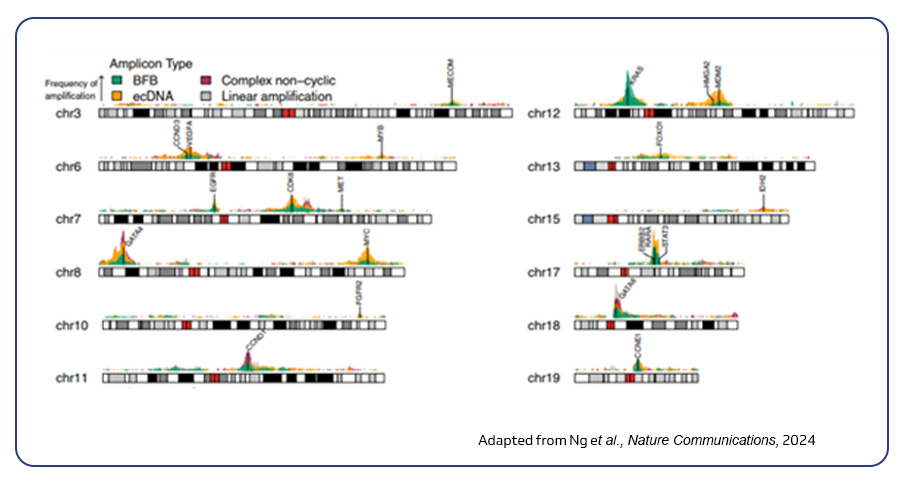
Single-cell TCR/BCR sequencing
Single-cell TCR/BCR sequencing technique is highly useful in understanding the immune system’s adaptive defenses by profiling T and B cell populations, uncovering rare immune cells crucial for fighting infections and tumors. The technique aids in analyzing unique variable regions within T and B cell receptors, enabling researchers to map antigen specificities and track immune cell development. Blanco-Heredia et al, Memorial Sloan Kettering Cancer Center, New York, used single-cell TCR sequencing to explore how the immune response interacts with tumor evolution in triple-negative breast cancer. The study found that as cancer progresses, the immune response weakens, marked by declining T cell diversity and the emergence of tumor escape mechanisms3.
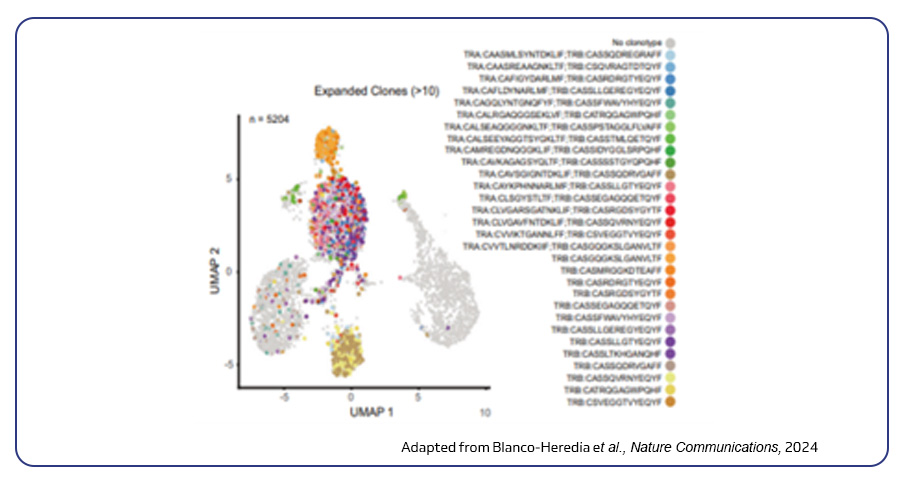
Single-cell ATAC-seq (scATAC-seq)
scATAC-seq decodes gene regulation within individual cells, identifying unique regulatory patterns and shedding light on cellular differentiation mechanisms. A study by Terekhanova et al. (2023) in Nature highlighted the impact of chromatin accessibility on cancer. Analyzing over 1 million cells from 225 tumor samples across 11 cancer types, the researchers found that changes in DNA accessibility can initiate, progress, and metastasize cancer. They identified both common and cancer-specific gene regulation patterns, including key pathways like TP53, hypoxia, and TNF signaling. The study also emphasized the role of estrogen response and epithelial-mesenchymal transition in metastasis, linking DNA accessibility to gene expression and cancer dynamics4.
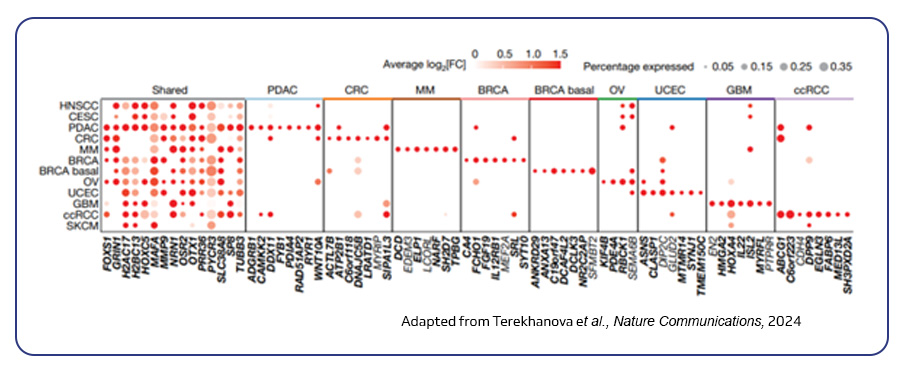
Single-cell multi-omics analysis
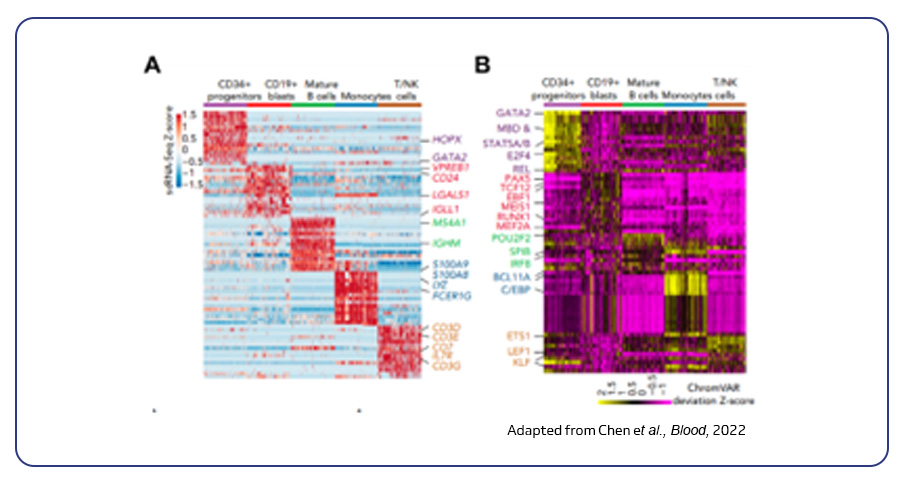
Single-cell multi-omics analysis integrates diverse data types beyond gene expression (scRNA-seq), including cell surface proteins, antigen receptors (TCR/BCR), and chromatin accessibility (scATAC-seq). CITE-seq further empowers this approach by simultaneously analyzing a vast number of cell surface proteins alongside gene expression in a single cell, offering a deeper understanding of cellular interactions and regulation. For example, a study using scRNA-seq and scATAC-seq on pediatric KMT2A-rearranged leukemia revealed key insights for targeted as well as immunotherapies5.
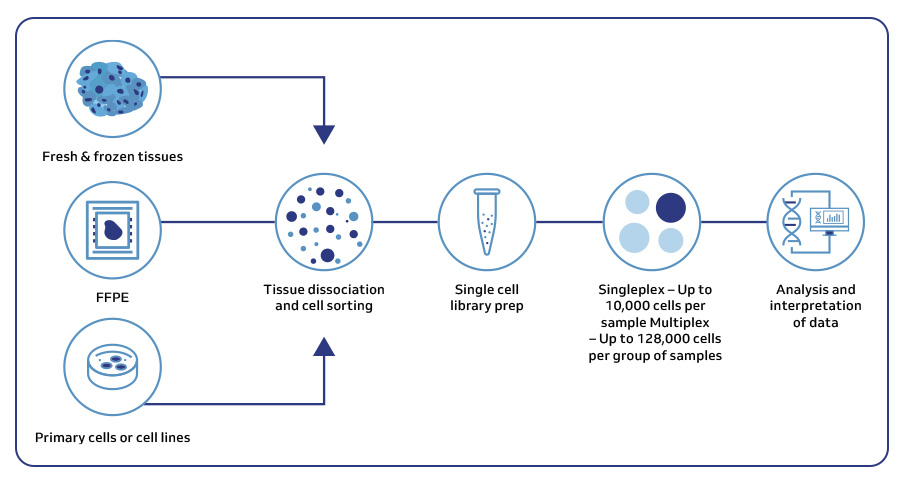
A seamless workflow for new discoveries
As a trusted 10x Genomics certified provider, MedGenome, offers end-to-end solutions tailored to your specific single-cell research needs. From sample preparation to data analysis, our expert team is there to support your project.
Extracting meaning from the data: Bioinformatics tools
At MedGenome, we have the expertise and the extensive computational infrastructure to provide a seamless experience from sample through insightful data analysis.
MedGenome offers a range of bioinformatics analysis options to meet your specific research needs:
- Standard analysis: We provide raw sequencing data (FASTQ files) and insights from Cell Ranger, including quality control metrics, gene expression levels, and heatmap visualizations for initial exploration using Loupe Browser software.
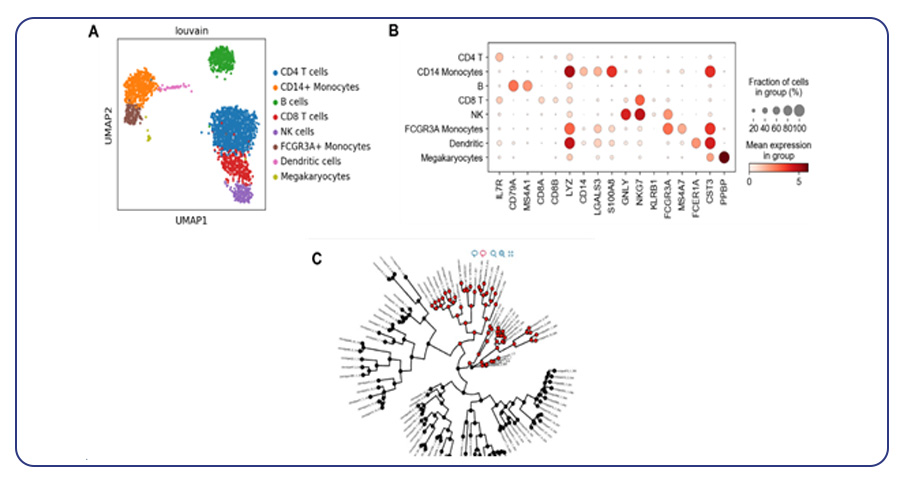
- Advanced analysis: MedGenome’s advanced analysis capabilities include all standard analysis deliverables along with advanced QC via Seurat and enhanced filtering of low-quality cells, contamination, and multiplets. The analysis also encompasses dimensionality reduction and clustering analysis on filtered data, interactive t-SNE plots with cluster information, and general cell type annotation. Additionally, MedGenome offers differential gene expression analysis for clusters and annotated cell types, pathway enrichment analysis, and group comparison.
- Specialized analysis: Tailor the analysis further with custom cell type annotations based on your unique markers. Uncover functional differences between cell types through differential gene expression analysis and map their interactions via cell-to-cell interactome analysis, providing a more comprehensive understanding of cellular behavior.
MedGenome, your single cell sequencing partner
MedGenome, a certified 10x Genomics partner, offers flexible and scalable single-cell sequencing solutions. We specialize in analyzing diverse samples—from fresh to cryopreserved and fixed specimens—using a range of advanced techniques. Our meticulous approach includes gentle tissue dissociation, viability checks, and precise cell sorting via FACS. Beyond processing, our expert bioinformatics team integrates samples, performs tailored analysis, and provides comprehensive, publication-ready reports. Researchers benefit from detailed metrics, plots, and raw data access for further exploration.
Unlock the full potential of your research with MedGenome’s cutting-edge single-cell sequencing solutions. Contact us today to learn how we can accelerate your scientific discoveries and drive groundbreaking insights in biology and medicine.
References
-
- Colella P, Sayana R, Suarez-Nieto MV, Sarno J, Nyame K, et al. CNS-wide repopulation by hematopoietic-derived microglia-like cells corrects progranulin deficiency in mice. Nat Commun 15, 5654 (2024).
- Ng AWT, McClurg DP, Wesley B, Zamani SA, Black E, et al. Disentangling oncogenic amplicons in esophageal adenocarcinoma. Nat Commun, 15, 4074 (2024).
- Blanco-Heredia, J., Souza, C.A., Trincado, J.L. et al. Converging and evolving immuno-genomic routes toward immune escape in breast cancer. Nat Commun 15, 1302 (2024).
- Terekhanova, N.V., Karpova, A., Liang, WW. et al. Epigenetic regulation during cancer transitions across 11 tumour types. Nature 623, 432–441 (2023).
-
Chen C, Yu W, Alikarami F, Qiu Q, Chen CH, Flournoy J, et al. Single-cell multiomics reveals increased plasticity, resistant populations, and stem-cell-like blasts in KMT2A-rearranged leukemia. Blood 139(14), 2198-2211 (2022).
#Single cell sequencing, #Single cell RNA sequencing, #Single cell DNA sequencing, #Single cell ATAC-seq, #Single cell TCR/BCR sequencing, #CITE-seq, #Single cell multiomics, #10x Genomics, #Single cell biomarker discovery, #Single cell clustering analysis, #Dimensionality reduction, #Personalized immunotherapy, #Single-cell tumor heterogeneity
 US
US IN
IN

