By Aditya Pai, VP Corporate and Business Development, MedGenome Inc.
World Cancer Day is a day to reflect and celebrate research victories, the battles that anyone with cancer fights, the search for new ways to detect cancer early and treat it as effectively as possible. Yet, cancer statistics remain sobering. Globally, there were an estimated 19.3 million new cancer cases and 10 million cancer deaths in 2020i. The number of people living with cancer is expected to grow by around 1 million every decade between 2010 and 2030ii.
Over the past decade, a better understanding of alterations in single driver genes, and genes involved in specific biological pathways have led to the development of better targeted therapies and more recently, immunotherapies. For example, tyrosine kinase inhibitors for non-small cell lung cancer or checkpoint inhibitors used as tissue agnostic immunotherapies have altered the treatment of many cancers. Yet, understanding the molecular mechanisms of various cancers is critical to develop new screening methods, e.g. non-invasive circulating tumor DNA ctDNA tests and novel therapies. The quest to improve overall survival rate in cancer with better and more targeted treatments has been abetted by significant investments by pharmaceutical and device companies as well as government funding. Precision medicine has also been aided by decreased costs of sequencing the genome which at large scale was cost prohibitive in the past. This has led to complex choices and decisions around whether to use a targeted gene panel or sequence the exome or whole genome. Various “omics” technologies have led to understanding the changes at the DNA level in a tumor to understanding the expression of various changes at the level of the transcriptome to understanding the tumor microenvironment using single-cell sequencing approaches. This multi-omic paradigm is gaining rapid traction in research. Yet, for a translational oncology researcher, a question I often get asked is “Should I sequence the entire genome or use a combination of whole exome sequencing and DNA sequencing or should I use a cancer panel?”
A translational oncology researcher is often looking for a broad set of results from a retrospective set of tumor samples stored as FFPE blocks (Formalin Fixed Paraffin Embedded). In this research discovery scenario, either of the above approaches have value, but it depends on factors such as the research hypothesis, time, research budget as well bioinformatics capabilities. For example, a Research Use Only (RUO) comprehensive gene panel like TSO 500 from Illumina includes 523 cancer related genes, includes (1) DNA variations like SNV’s, indels, copy number alterations (2) RNA gene fusion detection and (3) broad biomarker measurements like TMB that integrate many genomic loci across the genome. The TSO 500 panel has the flexibility in being used with solid tumor or ctDNA. The panel comes with added benefits of the bioinformatics pipeline included in the workflow. With more advanced bioinformatics, as we provide at MedGenome (see Figure 1, 2), a researcher could explore various hypothesis to better understand potential pathways and upstream and downstream cancer driver genes. Panels like TSO 500 are rigorously developed and designed to provide very robust, specific and sensitive (typically down to 5% variant allele frequency for most applications) detection of alterations in the genes of interest.
This same researcher could use a whole exome and RNA sequencing or whole genome sequencing approach. However, unlike the TSO 500 assay where bioinformatics analysis is included in the workflow, the researcher would have to separately perform bioinformatics analysis that can be time consuming. Yet, they may find this worthwhile if they are needing to explore the genome beyond what a panel of 523 genes would cover.
Key Takeaway
Sequencing and multi-omic approaches in oncology are quickly evolving and must be tailored to the primary questions that an oncology researcher is trying to answer. There are many approaches available.
At MedGenome, we can help you with your “omics” strategy. These include panel-based approaches that leverage comprehensive cancer genes, as well, whole genome / whole exome-based approaches, or more comprehensively single-cell RNA sequencing approaches.
In all these choices, cost is also an important consideration as is the ultimate use of these results which can range from exploratory research to a specific path for companion diagnostic development for a drug in clinical trials. Appropriate effort and detail to study design of a sequencing strategy and method is critical, as is the need for collaboration with high quality laboratories with validated assays.
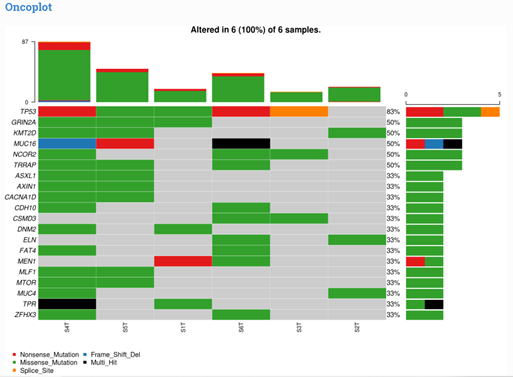
Figure 1: Oncoplot displaying the somatic landscape of the cohort for top (max 20) most frequently mutated genes. Each row represents a gene and each column represents a sample. Colored squares show mutated genes, while grey squares show non-mutated genes
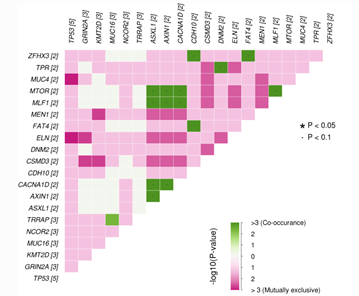
Figure 2: Somatic interaction plot shows mutually exclusive or co-occurring pair of genes displayed as a triangular matrix with top (max 20) most frequently mutated genes in the cohort. Green indicates tendency toward co-occurrence, whereas pink indicates tendency toward exclusiveness.
References
- I. https://www.uicc.org/news/globocan-2020-new-global-cancer-data
- II. https://www.macmillan.org.uk/_images/people-living-with-cancer_tcm9-283689.pdf
#cancer research, #targeted therapies, #tyrosine kinase inhibitors, #immunotherapies, #non-small cell lung cancer, #checkpoint inhibitors, #targeted gene panel, #translational oncology, #multi-omic approaches, #single-cell RNA sequencing
 US
US IN
IN

