By MedGenome Scientific Affairs
Head and neck squamous cell carcinoma (HNSCC) ranks among the most prevalent cancers worldwide. In the United States, it is estimated that 58,450 new cases will be diagnosed in 2024, primarily affecting the oral cavity and pharynx1. Incidence rates among males are highest in non-Hispanic White and American Indian/Alaska Native individuals, with lower rates observed in Hispanic and Asian/Pacific Islander populations. Among females, incidence rates are elevated in non-Hispanic White and Asian/Pacific Islander individuals, while being lowest in Hispanic and Black populations. Major risk factors for HNSCC include tobacco and alcohol use, as well as Human Papilloma Virus (HPV) infection. While tobacco-related HNSCC rates have declined over time, rising incidence rates, particularly among younger individuals, are attributed to HPV-related disease2.
Molecular pathogenesis of HNSCC
HNSCC, like other solid tumors, develops through genetic and epigenetic alterations, leading to various cancer phenotypes. Whole exome sequencing analyses of HNSCC specimens have identified mutations targeting key oncogenes and tumor suppressor pathways, such as p53, Rb/INK4/ARF, and Notch, which regulate cellular processes like proliferation, differentiation, and metastasis. Studies, including The Cancer Genome Atlas (TCGA) project, have categorized HNSCC based on genetic and expression patterns, revealing distinct subtypes with unique molecular features and clinical characteristics. Notably, mutations in genes like TP53 and CDKN2A are prevalent, while HPV+ tumors exhibit different mutation profiles, with frequent amplifications in PIK3CA and SOX2 genes. Furthermore, the Notch signaling pathway and PIK3CA alterations have been implicated in HNSCC progression and immune evasion3.
Tumor microenvironment of HNSCC
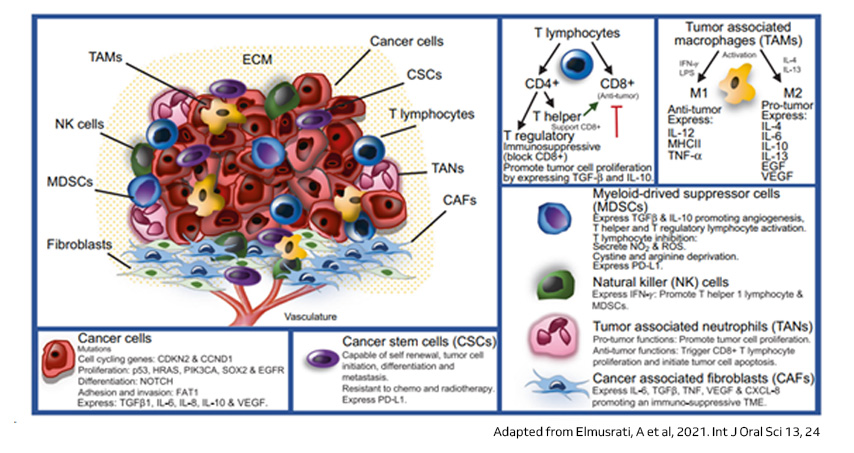
The tumor microenvironment (TME) in HNSCC and cancer generally comprises a diverse mix of cancer cells and nonmalignant components, including immune cells like T lymphocytes, tumor associated macrophages (TAMs), myeloid-derived suppressor cells (MDSCs), NK cells, tumor associated neutrophils (TANs), and Cancer associated fibroblasts (CAFs), along with fibroblasts, mesenchymal cells, and vascular endothelial cells. These nonmalignant cells play dual roles in tumor growth and dissemination. Understanding the immune landscape within the TME is crucial for assessing cancer progression and the efficacy of immunotherapy. Research on the TME in HNSCC highlights its impact on cancer behavior through complex cellular interactions mediated by growth factors and cytokines. Inflammatory processes within the TME worsen malignancy, with various immune cells exhibiting diverse functions affecting prognosis. Additionally, cytokines like TGFβ and IL-6 contribute to immunosuppression, influencing therapeutic responses, while hypoxia further promotes immune evasion and tumor progression3,4.
Single cell sequencing and its applications in HNSCC
Intra-tumoral heterogeneity (ITH) and plasticity present significant hurdles in translating cancer research into effective therapies due to the varied cellular compositions and dynamic functional states within tumors. Recent advances in single-cell sequencing have substantially improved the resolution of studies exploring ITH, the TME, and intra-tumoral cell-cell communication. This technology allows for the analysis of individual cells within the TME, revealing previously unseen diversity and dynamics. By identifying rare cell populations, characterizing cellular interactions, and discovering novel biomarkers and therapeutic targets, single-cell sequencing has the potential to revolutionize our understanding of HNSCC biology and pave the way for personalized diagnostic and therapeutic strategies. Some of the key highlights of single cell sequencing studies in HNSCC are provided below.
Insights into tumor composition and behavior
In one of the initial single cell sequencing studies on oral squamous cell carcinoma (OSCC), distinct non-malignant cell clusters were identified, including T-cells, macrophages, fibroblasts, and others, with T-cell subsets exhibiting variable sizes across patients. Conversely, malignant cells displayed patient-specific clustering, with only a few shared signatures among tumors, notably featuring a partial Epithelial-mesenchymal transition (EMT) program associated with advanced disease characteristics. Subsequent investigations involving a wider range of HNSCC subsites and HPV statuses corroborated these findings. These studies identified additional subclusters of fibroblasts and revealed patient-specific clustering patterns of malignant cells, with a correlation observed between these patterns and HPV status. Further exploration into cancer stem cells through scRNAseq highlighted metabolic program variations and extensive ITH across multiple tumor types, suggesting that transcriptional ITH reflects tissue heterogeneity5,6.
Decoding the complexity of TME
The immune system serves as a critical defense mechanism against malignant cells, leading to the development of immunotherapy as a prominent treatment strategy in cancer. However, despite its approval for HNSCC, immune checkpoint inhibitors only benefit a minority of patients. To understand the intricacies of the HNSCC TME, several studies have utilized single-cell RNA sequencing (scRNAseq) on sorted cell populations, including CD45+ hematopoietic cells or CD3+ T-cells. These studies often incorporate adjacent normal tissue, peripheral blood leukocytes, and non-tumorous tonsils for comparison. Analyses primarily focused on T-cells due to their pivotal role in antitumor immunity and immunotherapy. These studies revealed distinct T-cell subsets, such as CD8+ cells, which were found in higher proportions within tumor tissue compared to adjacent normal tissue. Additionally, investigations into CD4+ T-cells highlighted an increased presence of regulatory T-cells (Tregs) within tumors, indicative of an immunosuppressive tumor microenvironment. Furthermore, studies identified potential routes of cell-cell communication, particularly emphasizing the interaction between macrophages and T-cells mediated by PD-L1. Mouse models and scRNAseq data also showed how immune responses within tumors can vary, with T-cells multiplying in specific ways. This provided insights into tumor antigen recognition and how immune responses differ between patients. Explorations into the humoral arm of anti-tumor immunity and the contribution of natural killer (NK) cells underscored their potential therapeutic implications in HNSCC5,6.
Conclusions
HNSCC presents a complex interplay between genetics, the tumor microenvironment, and the immune system. Understanding these factors is crucial for developing effective therapeutic strategies. Single-cell sequencing has emerged as a powerful tool, shedding light on the intricate cellular diversity within HNSCC tumors and the dynamic interactions within the TME. These insights not only hold promise for personalized medicine in HNSCC but also contribute significantly to our overall understanding of cancer biology, paving the way for advancements in cancer research across different tumor types.
MedGenome solutions
As a 10x certified service provider and an early pioneer in single-cell genomic sequencing, MedGenome offers comprehensive support throughout your research journey, from experimental design to publication. Our expertise spans selecting the most suitable single-cell workflow, processing diverse sample types with efficiency and accuracy, and delivering timely results. With custom visualizations, tailored analysis workflows, and seamless integration of external data, we ensure your research is publication-ready. Our proprietary algorithm, OncoPeptTUMETM utilizes RNA-seq data to create high-resolution maps of the tumor microenvironment based on specific cell type gene signatures.
For any queries or additional details, please reach out to our expert scientific team at research@medgenome.com.
To know more about our unique cancer genomics solutions and services please click on the following links: Whole genome and whole exome sequencing, RNA Sequencing, Single cell sequencing, Immune profiling and Epigenetic profiling
References
-
- Siegel RL, Giaquinto AN, Jemal A. Cancer statistics 2024 (2023). CA Cancer J Clin. 2024. 74(1):12-49.
- Barsouk A, Aluru JS, Rawla P, Saginala K, Barsouk A. Epidemiology, Risk Factors, and Prevention of Head and Neck Squamous Cell Carcinoma (2023). Med Sci (Basel). 13;11(2):42
- Elmusrati, A., Wang, J. & Wang, CY. Tumor microenvironment and immune evasion in head and neck squamous cell carcinoma (2021). Int J Oral Sci 13, 24.
- Ruffin AT, Li H, Vujanovic L, Zandberg DP, Ferris RL, Bruno TC. Improving head and neck cancer therapies by immunomodulation of the tumour microenvironment. Nat Rev Cancer. 23(3):173-188.
- Qi Z, Barrett T, Parikh AS, Tirosh I, Puram SV. Single-cell sequencing and its applications in head and neck cancer (2019). Oral Oncol. 99:104441.
- Heller G, Fuereder T, Grandits AM, Wieser R. New perspectives on biology, disease progression, and therapy response of head and neck cancer gained from single cell RNA sequencing and spatial transcriptomics (2023) Oncol Res. 32(1):1-17.
#Head and Neck cancer, #Oral squamous cell carcinoma, #Tumor microenvironment, #Single cell sequencing, #Immunotherapy, #Immune checkpoint inhibitors, #Intra-tumoral heterogeneity, #Epithelial-mesenchymal transition, #Human papilloma virus
 US
US IN
IN

