TCR/BCR Sequencing from MedGenome
The immune repertoire is the complete collection of clonotypes for any particular receptor chain. Studying the immune repertoire can reveal a lot about a person’s current health while also providing a record of their immune challenges and changes over time. Each T and B cell produce only a single receptor clonotype. Since the number of cells in the adaptive immune system is finite, when one clonotype expands, others are displaced. MedGenome offers several solutions for immune profiling, including bulk and single-cell TCR and BCR sequencing.
MedGenome TCR Sequencing Work Flow
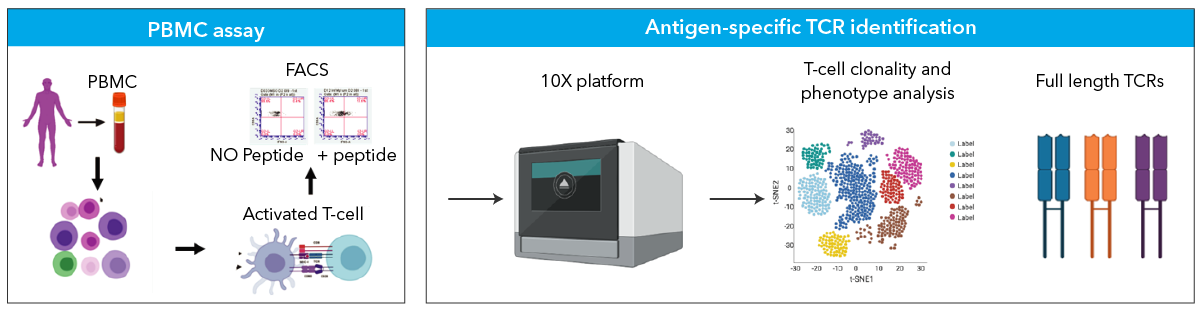
Bulk TCR / BCR
Detecting changes to the TCR or BCR repertoire is a rapidly developing strategy to discover biomarkers of disease progression or response to a vaccine or immunotherapy. MedGenome uses a RNA-based method for both TCR and BCR sequencing. These methods start with total RNA and utilize Takara SMART (Switching Mechanism at the 5’ end of RNA Template) technology to generate cDNA. Full length variable regions are then amplified using a nested PCR. Finally, a library is constructed. Using RNA, rather than DNA, as input allows for greater sensitivity and detection of low abundance TCR/BCR variants from small sample inputs. Additionally, UMIs are utilized to reduce PCR/sequencing errors in the data.
Single-Cell TCR / BCR
MedGenome utilizes the latest technology by 10x Genomics to capture and sequence thousands of cells per sample. Single-cell immune profiling generates paired, full-length immunoglobulin sequences, including isotypes, as well as TCR alpha/beta sequences. This can be combined with gene expression or cell-surface protein data for multi-omic analysis. Single-cell immune profiling is a great way to accelerate biomarker discovery and therapeutics research.
Features and Benefits
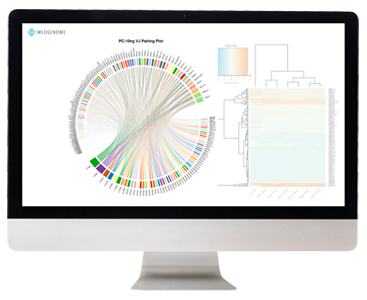
Analysis
Our data analysis starts with the generation of a Multi-QC Report. This is a modular tool to aggregate results from bioinformatics analyses across many samples. This report provides deep insight on overall data QC, alignment statistics, and a FastQC report. These data represent the overall success of the library prep and sequencing.
Reads are aligned to the transcriptome and expression values are reported in FPKM (Fragment Per Kilobase per Million). Groups of samples can then be compared to detect differentially expressed genes. We generate publication-ready plots, such as heat maps and volcano plots, to visualize significant trends in the data. We also offer pathway analysis and tumor microenvironment analysis for deeper understanding of these data.
Company
Resources
Research
- Tumor Microenvironment Solutions
- Immune Profiling Services
- NGS Services
- BioFX Bioinformatics Services
- Antibody Discovery Services
Diagnostics
Contact

