Single Cell Sequencing Services by MedGenome
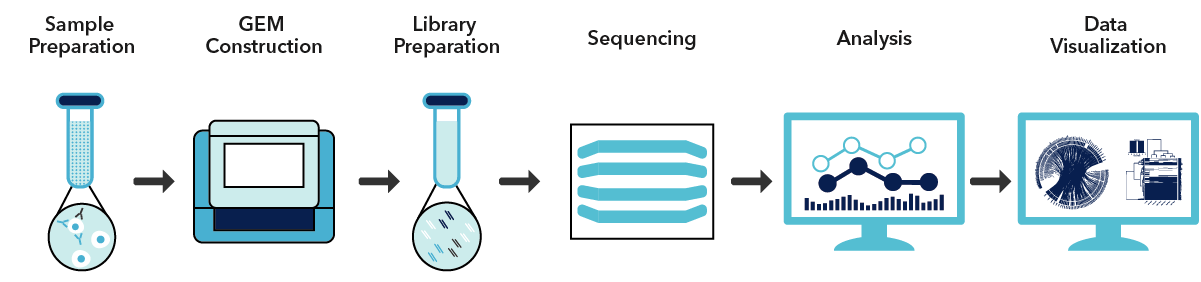
Cell populations are often heterogeneous with diverse characteristics. Single-cell RNA sequencing aims to uncover the transcriptome diversity in these samples. This is possible by using microfluidics to isolate single cells and molecular barcoding to label the transcripts associated with those cells. MedGenome utilizes the latest technology by 10x Genomics to capture and sequence thousands of cells per sample. This platform provides a cost-effective method for analyzing heterogeneous cell populations.
Single-Cell Gene Expression Profiling
This basic approach to single-cell sequencing allows users to analyze transcriptome heterogeneity at the single-cell level. MedGenome offers optimized workflows for 5’ and 3’ gene expression libraries. These solutions can be used to uncover cellular differences that are usually masked by bulk RNA-sequencing experiments.
Single-Nuclei Sequencing
Depending on the project type, single nuclei may be a better fit as sample input. This is common when studying tissue samples which it is difficult to extract intact cells, such as brain tissue or flash frozen tissue. MedGenome has extensive experience conducting nuclei isolation from tissues to generate sequencing libraries.
Single-Cell Immune Profiling
This method allows scientists to examine the immune profile of T-cells and B-cells in their samples. Libraries contain information for analysis of full-length V(D)J sequences for paired B-cell and T-cell receptors. This can be combined with single-cell gene expression profiling for a multiomic approach to single-cell sequencing.
Single-Cell Epigenetics
Identify chromatin accessibility in the epigenome by profiling thousands of single cells in parallel. Discover how chromatin structure and DNA-binding proteins regulate gene expression in various states and cellular processes. MedGenome offers solutions for stand alone ATAC-seq, and also offers combined ATAC-seq and gene expression from the same cells.
CITE-Seq & Cell Multiplexing
CITE-seq uses DNA-barcoded antibodies to convert detection of cell-surface proteins into a quantitative sequenceable readout. Antibody-bound oligos from BioLegend act as synthetic transcripts that are captured during single-cell sequencing. This allows for the profiling of cell surface markers alongside unbiased transcriptome analysis. This same strategy can be used for cell multi-plexing. Oligo-tagged antibodies can be used to differently label several samples. These samples can then be combined into a single library, sequenced, and then the data can be deconvoluted based on these unique tags.
Single-Cell Techniques
| Single Nuclei | Single Cell | Immune Profiling | CITE-Seq | Hashtag Antibodies | |
| 3' Gene Expression |  |  |  |  | |
| 5' Gene Expression |  |  |  |  | |
| ATAC-Seq |  | ||||
| Multiome (Gene Expression + ATAC-Seq) |  |
Tissue Source
-
 PBMC
PBMC -
 Liver
Liver -
 Brain
Brain -
 Heart
Heart -
 Lung
Lung -
 Kidney
Kidney -
 Cell culture (live/fresh cells)
Cell culture (live/fresh cells)
Features & Benefits
Analysis
Our data analysis starts with the generation of a Multi-QC Report. This is a modular tool to aggregate results from bioinformatics analyses across many samples. This report provides deep insight on overall data QC, alignment statistics, and a FastQC report. These data represent the overall success of the library prep and sequencing.
Reads are aligned to the transcriptome and expression values are reported in FPKM (Fragment Per Kilobase per Million). Groups of samples can then be compared to detect differentially expressed genes. We generate publication-ready plots, such as heat maps and volcano plots, to visualize significant trends in the data. We also offer pathway analysis and tumor microenvironment analysis for deeper understanding of these data.
Company
Resources
Research
- Tumor Microenvironment Solutions
- Immune Profiling Services
- NGS Services
- BioFX Bioinformatics Services
- Antibody Discovery Services
Diagnostics
Contact

